Improving The Treatment Of Acute Lymphoblastic Leukemia Using Graph Analytics With AI And Machine Learning
- Blog >
- Improving The Treatment Of Acute Lymphoblastic Leukemia Using Graph Analytics With AI And Machine Learning
This is an abbreviated version of a presentation by Jesper Vang, Technical University of Denmark, during the Graph + AI Summit 2021 conference
Our group at the Technical University of Denmark is centered on translational bioinformatics. Our main objective is to improve the diagnostics, treatment, and cure rates for children with cancer. We focus mainly on developing storage interpretative models and methods, and also optimizing the transformation of increasingly large biomedical and genomic data that’s been generated.
Currently, we are building a biobank and database for childhood cancer research that includes samples, both retrospective, and prospective. While we’re generating the data, we also carry out extensive analysis on germline and tumor DNA, as well as transcriptomics, to gain insight into cancer, somatic mutations, tumor biology, and predisposition syndromes.
By linking together various other data points about the patient’s life, illness, and treatment, clinical personnel can understand to a much greater extent why children get cancer and provide earlier diagnosis and far more effective treatment.
Our long-term goal is to create one of the most respected, multidisciplinary research groups within childhood cancer. To achieve that we of course need great partners, researchers and also a great data infrastructure.
How are we using graph analytics to help? Well, as you can imagine, we have a lot of data. We feel limited by our relational database. Why? Although a relational database handles close relationships between data easily, they are more challenging when investigating distant relationships.
When we started building our relational database, we built it with questions in mind, but the problem with research is that you never really know what the perfect question is in the beginning. With a graph database, it’s possible to answer random questions, which is typical for us as a hospital or doctor comes up with a new question and expects the answer to be somewhere in the data.
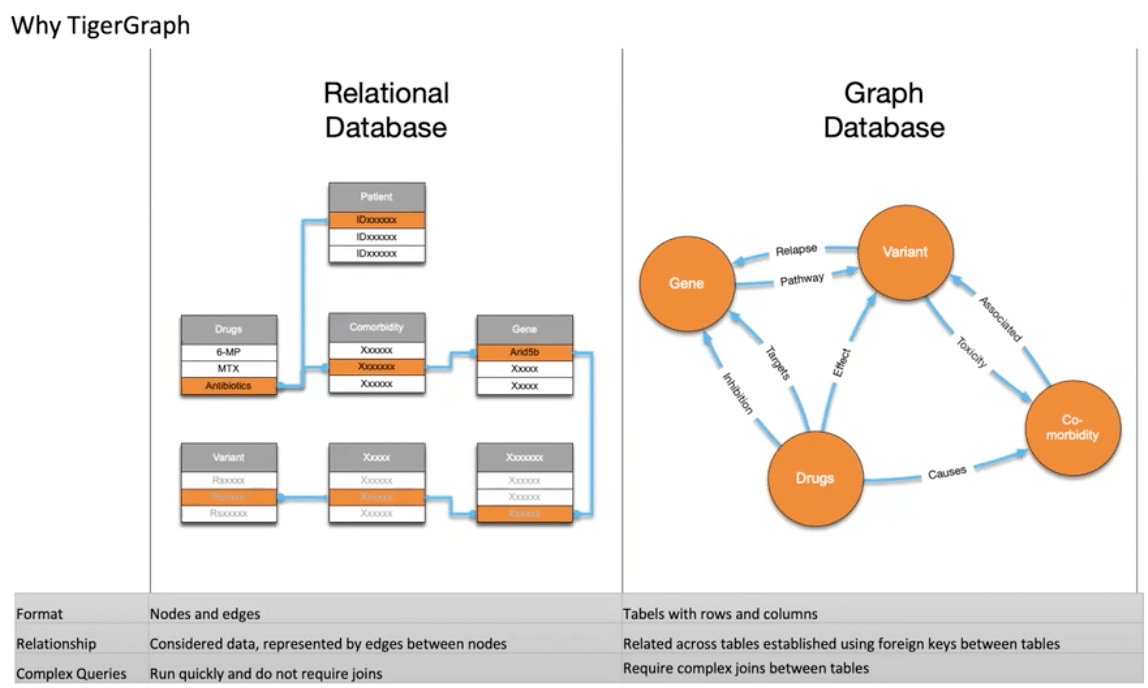
For example, let’s say you want to determine how many patients, with a specific variant, were treated with a specific drug, but experienced some type of complication. In a relational database, the database administrator would have to sort of predict that these questions would be asked at some point. Otherwise, it would be really difficult to retrieve that information.
With a graph database, however, it’s possible to answer random questions—and that’s necessary for a medical environment where a doctor could come up with a question and needs an answer from the data. By using a graph we can answer all the questions, as long as there is a relationship between the data, and we can add more data and relationships as the information emerges whilst maintaining the same performance.
Analyzing relationships between nodes representing patients, and nodes representing genetic variations, is as simple as finding the connecting nodes. And we do not have to redefine the database every time we add new annotations. Why? This is because the graph database is dynamically extensible to accommodate new sources of information so we do not have to redefine the database every time we add data sources.
I hope that this was informative, and am looking forward to showcasing the results of our research at the next Graph + AI summit. I would like to thank TigerGraph for being very supportive, something that we really appreciate. And, finally, I would like to thank the team at the university, who is doing some truly excellent work in cancer research.
You can listen to the full presentation here.